Band structure of Ni
From phys824
Input files
- Save two files below as *.py files.
band.py
from gpaw import GPAW, FermiDirac
from ase import Atoms
from ase.io import read, write
from gpaw import GPAW, PoissonSolver, MixerSum
from ase.structure import bulk
# -------------------------------------------------------------
# Bulk configuration
# -------------------------------------------------------------
a = 3.5249
atoms = bulk('Ni', 'fcc', a=a)
atoms.center()
write('system.traj', atoms)
for a in atoms:
if a.symbol == 'Ni':
a.magmom = 0.6
# Make self-consistent calculation and save results
calc = GPAW(h=0.18,
mode='lcao',
xc='PBE',
basis='dzp',
kpts=(8,8,8),
occupations=FermiDirac(width=0.05, maxiter=2000),
mixer=MixerSum(beta=0.010, nmaxold=8, weight=100.0),
poissonsolver=PoissonSolver(eps=1e-12),
txt='band_sc.txt')
atoms.set_calculator(calc)
atoms.get_potential_energy()
calc.write('band_sc.gpw')
# Calculate band structure along Gamma-X
from ase.dft.kpoints import ibz_points, get_bandpath
points = ibz_points['fcc']
G = points['Gamma']
X = points['X']
kpts, x, X = get_bandpath([G, X], atoms.cell, 80)
calc = GPAW('band_sc.gpw',
mode='lcao',
xc='PBE',
basis='dzp',
kpts=kpts,
txt='band_harris.txt',
fixdensity=True,
parallel={'domain': 1},
usesymm=None,
convergence={'bands': 'all'})
if calc.input_parameters['mode'] == 'lcao':
calc.scf.reset()
calc.get_potential_energy()
calc.write('band_harris.gpw')
find_band_as_dat.py
from ase.lattice import bulk
from gpaw import GPAW
calc = GPAW('band_harris', txt=None)
import numpy as np
eps_skn = np.array([[calc.get_eigenvalues(k,s)
for k in range(80)]
for s in range(2)]) - 10.75103
for n in range(10):
for k in range(80):
print k, eps_skn[0, k, n], eps_skn[1, k, n]
print
Job submission script
- Save the following into a file jobscript within your working directory
#$ -N band
#$ -o out.log
#$ -e band.err
#$ -pe openmpi 16
#$ -l h_rt=3:00:00
#
source /opt/shared/valet/docs/valet.sh
vpkg_rollback all
vpkg_require numpy/1.6.1-2.7
vpkg_require mkl/10.3.8-64bit
vpkg_require openmpi/1.6.1-intel64
#
export PYTHONPATH=/lustre/scratch/ksaha/gpaw/ase:$PYTHONPATH
export PATH=/lustre/scratch/ksaha/gpaw/ase/tools:$PATH
export GPAW_SETUP_PATH=/lustre/scratch/ksaha/gpaw/gpaw-setups-0.8.7929
export PYTHONPATH=/lustre/scratch/ksaha/gpaw/gpaw:${PYTHONPATH}
export PYTHONPATH=/lustre/scratch/ksaha/gpaw/gpaw/build/lib.linux-x86_64-2.7:${PYTHONPATH}
export PATH=/lustre/scratch/ksaha/gpaw/gpaw/build/bin.linux-x86_64-2.7:/lustre/scratch/ksaha/gpaw/gpaw/tools:${PATH}
#
#
cd /home/1076/FCC-Ni_bandstructure/Wiki
#
mpirun /lustre/scratch/ksaha/gpaw/gpaw/build/bin.linux-x86_64-2.7/gpaw-python band.py
#mpirun /lustre/scratch/ksaha/gpaw/gpaw/build/bin.linux-x86_64-2.7/gpaw-python find_band_as_dat.py
Running the job script
- To run band.py execute:
qsub jobscript
- To run find_band_as_dat.py execute:
- 1. modify the 4th line of the jobscript "#$ -pe openmpi 16" to "#$ -pe openmpi 1" -- this means we will be running in a single core.
- 2. comment the second last line and uncomment the very last line of the jobscript
- 3. then submit job: qsub jobscript
- 4. bands should be generated into "out.log" file
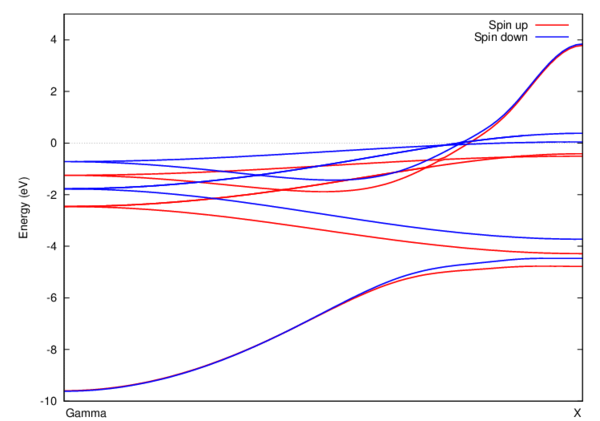
Test results
- Once you successfully finish running both jobs, you should be able to reproduce this figure (using gnuplot on Mills or transfer data to your Windows PC via sftp and use Origin):
gnuplot> plot "result.dat" using 1:2 with lines title "Spin up", "result.dat" using 1:3 with lines title "Spin down"
 |